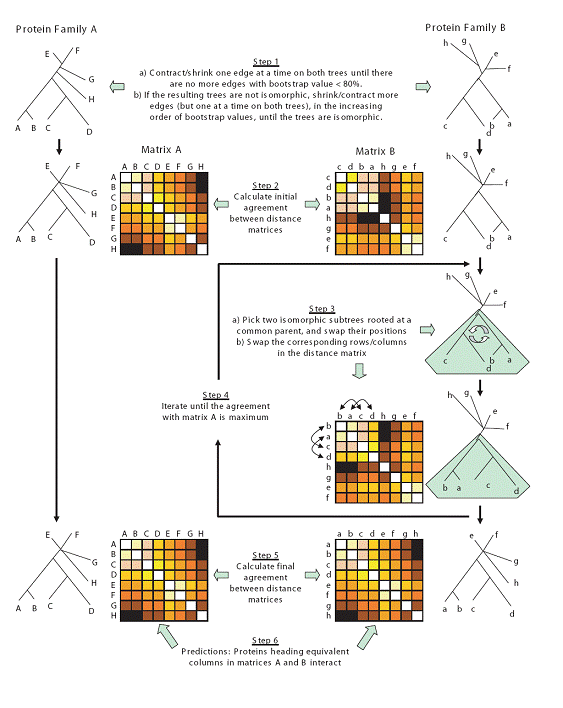
Motivation : Uncovering the protein-protein interaction network is a fundamental step in the quest to understand the molecular machinery of a cell. This motivates the search for efficient computational methods for predicting such interactions. Among the available predictors are those that are based on the co-evolution hypothesis "evolutionary trees of protein families (that are known to interact) are expected to have similar topologies". Many of these methods are limited by the fact that they can handle only a small number of protein sequences. Also, details on evolutionary tree topology are missing as they use similarity matrices in lieu of the trees. Results: We introduce MORPH, a new algorithm for predicting protein interaction partners between members of two protein families that are known to interact. Our approach can also be seen as a new method for searching the best superposition of the corresponding evolutionary trees based on tree automorphism group. We discuss relevant facts related to the predictability of protein-protein interaction based on their co-evolution. When compared with related computational approaches, our method reduces the search space by approximately 3 x 10(5)-fold and at the same time increases the accuracy of predicting correct binding partners.
|